DNA Hybridization on Surfaces - Learn More
Bulk hybridization of DNA oligonucleotides is understood as a robust phenomenon. This phenomenon was used to design DNA microarrays, devices that hybridize sample DNA to a surface to perform thousands of genomic assays with a single experiment. The resulting devices, however, do not have the consistent reproducibility seen in experiments that allowed oligonucleotides to hybridize in the bulk. This study utilizes molecular level simulations to characterize the hybridization of DNA oligonucleotides in bulk.
Protein-Surface Interactions - Learn More
Folding Cooperativity With Multiple Proteins - Learn More
We are exploring the effects of simulating multiple proteins of the same kind to better approximate bulk condtions. We are especially interested in multi-state proteins where some transitions become more cooperative, and some less cooperative. We hope to use this to help explain discrepancies between simulation and experiment.
Simulating HPLC - Learn More
How Mutations Affect Protein Function
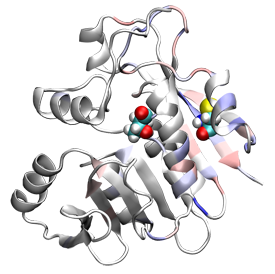
By tracking the stress of each atom in a protein, we can follow how a mutation changes these stresses for each atom. This will help to predict how the mutation affects the overall function of the protein. In the figure, two mutations are show as colored atoms, while the red and blue tinting shows changes in stress.